

Amyloid fibrils
Amyloid fibril is a needle-like aggregate of proteins and peptides. Irrespective of precursor, the fibril exhibits similar morphology, i.e., no branch, typically ~μm in length, and ~10 nm in thickness. The fibrils of more than 20 proteins are related to human diseases. The fibril of À2-microglobulin (À2m) is found in patients undergoing long-term dialysis and recognized as the origin of dialysis-related amyloidosis.
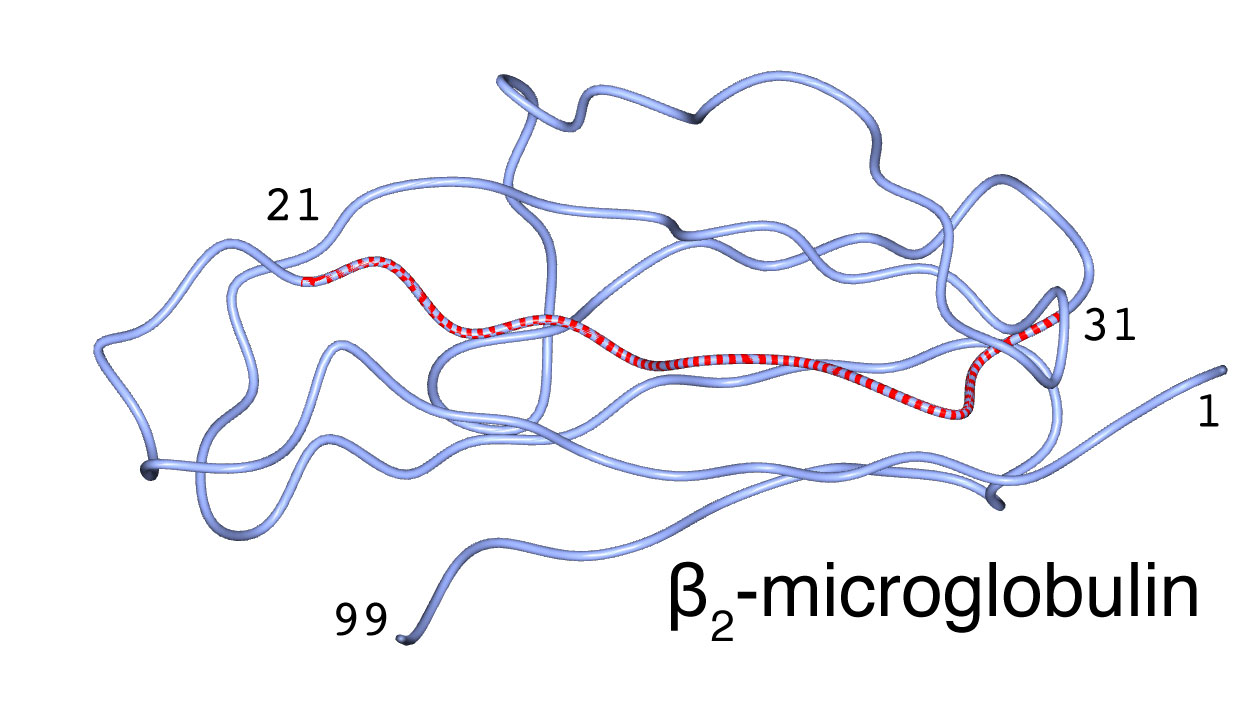
Core structure of À2m amyloid fibril
The À-sheet structure predominates in the fibril, and the À-strand runs perpendicular to the fibril long axis. Loop, bulge, turn also exist therein. In order to make the analysis simpler, we used peptide fragments [21NFLNCYVSG29 (À2m21-29) and 21NFLNCYVSGFH31 (À2m21-31)] that have partial sequence of À2m and that have self-fibrilation propensity, and analyzed the molecular structure of the peptide fibrils by means of IR linear dichroism spectroscopy, the 13C isotope-edited IR spectroscopy, vacuum-UV circular dichroism spectroscopy, and low-frequency Raman spectroscopy. These experiments cleared detailed molecular structure of the amyloid fibril of À2m21-31.
[1] H.Hiramatsu, Y.Goto, H.Naiki, T.Kitagawa, J. Am. Chem. Soc. 2004, 126, 3008-3009.
[2] H.Hiramatsu, Y.Goto, H.Naiki, T.Kitagawa, J. Am. Chem. Soc. 2005, 127, 7988-7989.
[3] H.Hiramatsu, T.Kitagawa, Biochimica et Biophysica Acta 2005, 1753, 100-107.
[4] K.Matsuo, H.Hiramatsu, K.Gekko, H.Namatame, M.Taniguchi, R.Woody, J. Phys. Chem. B 2014, 118, 2785-2795.
[5] S.Shigeto, C.-F.Chang, H.Hiramatsu, J. Phys. Chem. B, 2017, 121, 490-496.
Origin of difference in morphology
Besides the fibril having the needle-like morphology, À2m forms "worm-like" aggregates in the presence of high salt concentration. In order to get insight into an origin of the morphological difference, we applied the IR and Raman (and vacuum-UV circular dichroism) spectroscopy to the needle-like and worm-like samples. From the analysis of the COOH and COO- bands in the spectra, we found that the morphological difference was accompanied by different ionization states of the carboxyl groups. It suggests the difference in the formation of hydrogen bonds (salt bridges) that the carboxyl groups take part in between the two kinds of the aggregates.
[5] H.Hiramatsu, M.Lu, K.Matsuo, K.Gekko, Y.Goto, T.Kitagawa, Biochemistry, 2010, 49, 742-751.
13C isotope edition for structural analysis
The C=O stretching of the peptide main chain gives a strong IR absorption (Amide I, AmI) band, and the band feature of the AmI band is known to give information about the secondary structure of the peptides and proteins. This procedure was applied to the structure analysis of the amyloid fibril, and the fraction of the secondary structures (i.e., the À-sheet and irregular structures) has been determined. In order to get insight into the peptide structure in more detail, we performed the 13C isotope labeling on the peptide main chain and analyzed the peak position of the AmI band of the 13C-labeled residue. The secondary structure of each residue was successfully determined. The structural information on "each residue" is sufficiently precise to discuss several problems.
[6] H.Hiramatsu, Y.Goto, H.Naiki, T.Kitagawa, J. Am. Chem. Soc. 2005, 127, 7988-7989.
[7] M.Lu, H.Hiramatsu, Y.Goto, T.Kitagawa, J. Mol. Biol. 2006, 362, 355-364.
[8] H.Hiramatsu, M.Lu, Y.Goto, T.Kitagawa, Bull. Chem. Soc. Jpn 2010, 83, 495-504.
À2m21-29 as a model peptide of the parallel/anti-parallel sheet structures
We found that a nine-residue peptide having the 21-29 partial sequence of À2-microglobulin (À2m21-29) formed the parallel À-sheet (PÀ) above pH 7.8 and the anti-parallel À-sheet (APÀ) below pH 7.6; the structure was confirmed by executing the IR linear dichroism and 13C isotope labeling[8].
The strength of the intermolecular interaction of the structures having the distinct stacking was compared by measuring the low-frequency Raman spectrum, and the stronger interaction in PÀ was concluded from the higher vibrational frequency[5]. Form detailed analysis of the pH dependence, we showed that the two À-sheet structures does not mix (i.e., the mixed À-sheet never forms), and explained this observation by taking account of the difference of the dihedral angles of the peptide main chain in the À-strand[9].
[9] W.-H.Tseng, S.-H.Chen, H.Hiramatsu, Sci. Rep. 2020, 10, 22199.